
Course
BIOINFORMATICS


Course
BIOINFORMATICS
|
|
Teachers: Alessio Bechini (instructor) CFU: 6 Code: 688II  Bioinformatics is an interdisciplinary field where both conceptual and actual tools typical of Computer Science are used to tackle a wide variety of issues in Molecular Biology. In particular, Bioinformatics adopts computational approaches in the management and analysis of biological data. This class provides an introduction to some topics in
Bioinformatics, emphasizing concepts and tools to master the inherent
complexity of the target systems. An operative approach is pursued: in
problem solving, a combined employment of multiple software
applications and a scripting language is proposed. The first part of
the class is thus devoted to a thorough teaching of the Python
language. Discussed topics include sequence analysis, mutation
investigation and molecular evolution, interaction with data banks,
molecular modeling, protein structural characterization, and molecular
dynamics. Whenever needed, notions of Computers Science and Maths are
introduced, e.g. computational complexity and integration schemes. The course is aimed at properly developing the skills of
future Biomedical Engineers in the Computer Science field. Students
will acquire the expertise required to participate in design,
implementation and integration of heterogeneous software systems in the
area of Bioinformatics and Computational Biology. Prerequisites: knowledge of Computer Science basics, and mastering of at least one programming language; fundamental notions of Physics, Chemistry, and Molecular Biology. -
Available only in the italian version. -
Available only in the italian version. A detailed description is available via the record of lessons (see aside). Hereafter, a cathegorized list of topics can be interactively explored. -
Available only in the italian version. -
Available only in the italian version. A list of software tools among the most popular ones in the proteomics field can be found on the ExPASy site. Open positions for jobs in the field of Bioinformatics and Computational Biology can be found in dedicated pages from BITS, ISBC, and bioinformatics.org -
Further information available only in the italian version. Notes
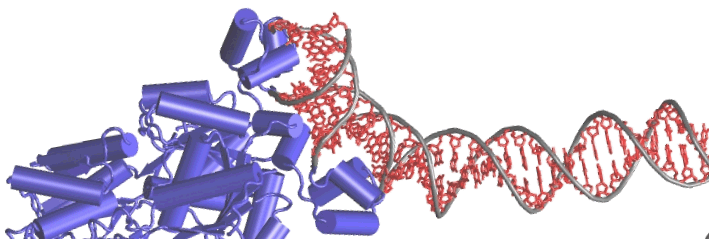
The picture on the right side of the heading illustrates a particular of protein "lac repressor" (a genic regulator in E.Coli) as it binds to a DNA strand (TCB - Univ. of Illinois at Urbana-Champain) The background picture represents a portion of an insuline model, as visualized in the VMD application. The picture in the "description" section represents a "tag-cloud" with popular terms in bioinformatics papers. |